Atom lines from
1W1X,
4FVK,
4K3Y,
1A4G, and
3H72
were used above to provide a visual example of dist/deviation use.
DIST and DEVIATION Tutorial: Making a population standard deviation matrix for the interatomic distances of 22 specific main chain oxygen atoms in selected neuraminidase proteins
Overview
Instructions
Summary
(WEB PAGE PDF)
Tutorial Overview
This tutorial shows how to easily create a population standard deviation matrix for the interatomic distances within separate molecules. This tutorial will first show how to use dist to create distance matrices from selected lists of atoms (in PDB format). This tutorial will then show how to use deviation to create a population standard deviation matrix from these distance matrices.
- dist creates a distance matrix from a points list.
There are options for handling input and output labels and specifying output format.
dist can optionally parse a PDB file, and has options for specifying user defined output atom labels.
- deviation reads in multiple files and writes out the population standard deviation
of numerical values that match string token positions across all input files.
The dist and deviation Overview Pages describes dist and deviation command line syntax.
Specific neuraminidases (N6 (1W1X), N10 (4FVK), N11 (4K3Y), influenza B (1A4G), and streptococcus (3H72)) contain distributed atoms that have common spatial occupancy, i.e. atoms that occupy similar relative positions in space.
dx.doi.org/10.1371/journal.pone.0117499
(16 MB PDF – LOCAL COPY) (3 MB PDF – LOCAL COPY)
This tutorial will create a population standard deviation matrix for the interatomic distances of 22 selected main chain oxygen atoms from these neuraminidase proteins.
Instructions
- Request and install dist and deviation
(if you have not already done so).
https://weiningerworks.com/contact.html - Download, (from weiningerworks.com by using the links below) five files that are subsets of PDB structure files from rcsb.org:
1W1Xatoms.pdb
4FVKatoms.pdb
4K3Yatoms.pdb
1A4Gatoms.pdb
3H72atoms.pdb
These files should have a similar format to the following (and 1W1Xatoms.pdb should have identical content):
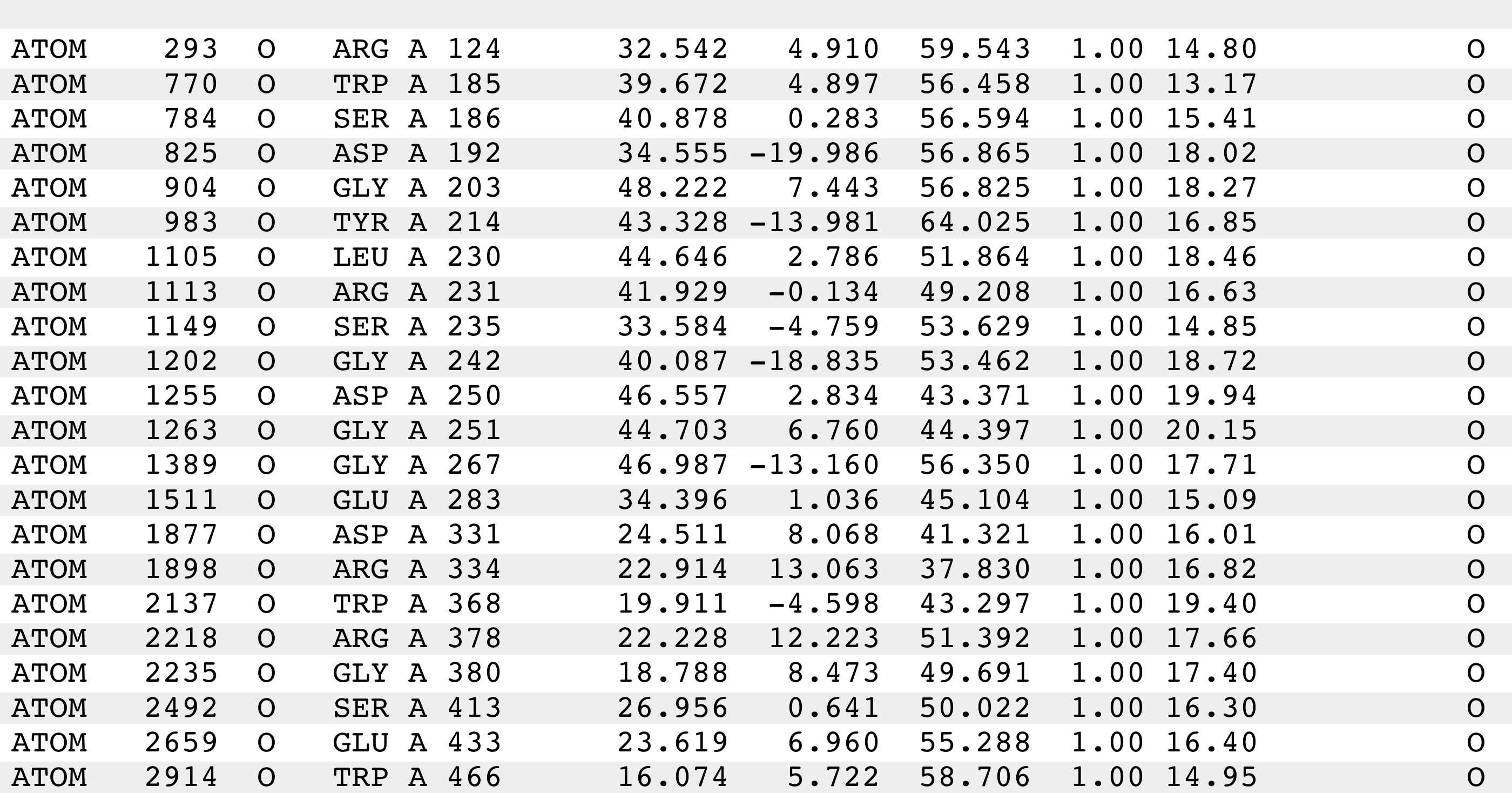
- Run dist to create distance matrices for each of these lists of atoms.
dist will output a single distance matrix file (1W1Xatoms.dist, 4FVKatoms.dist, 4K3Yatoms.dist, 1A4Gatoms.dist, 3H72atoms.dist in the following) for each dist execution. (Option '-i' specifies the input list filename, option '-o' specifies the output distance matrix filename, and option '-a' specifies that the input list is in PDB format.)
dist -i 1W1Xatoms.pdb -o 1W1Xatoms.dist -a
dist -i 4FVKatoms.pdb -o 4FVKatoms.dist -a
dist -i 4K3Yatoms.pdb -o 4K3Yatoms.dist -a
dist -i 1A4Gatoms.pdb -o 1A4Gatoms.dist -a
dist -i 3H72atoms.pdb -o 3H72atoms.dist -a
These files should have a similar format to the following (and 1W1Xatoms.dist should have identical content):

- Run deviation to create a single population standard deviation matrix, named 'neuraminidases.deviation', from the newly created five distance matrices.
(Option '-o' specifies the output population standard deviation matrix filename, the input files are listed at the end of the deviation command.)
deviation -o neuraminidases.deviation 1W1Xatoms.dist 4FVKatoms.dist 4K3Yatoms.dist 1A4Gatoms.dist 3H72atoms.dist
The newly created file 'neuraminidases.deviation' should have identical content to the following:
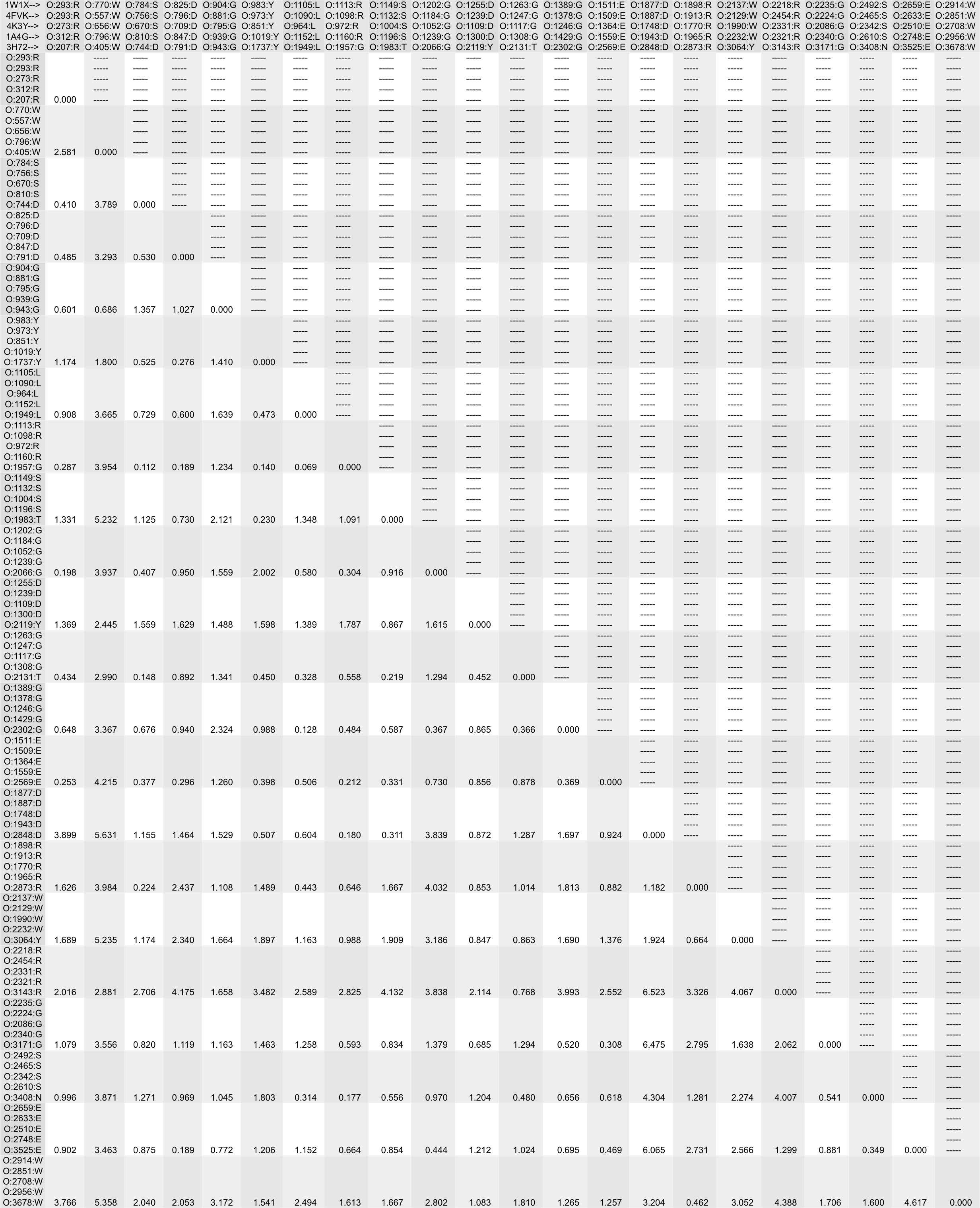
Tutorial Summary
dist created distance matrices from selected lists of atoms (in PDB format). deviation created a population standard deviation matrix from these distance matrices.
If you are interested in identifying atoms in molecules that have common spatial occupancy (i.e. atoms that occupy similar relative positions), check out the Weininger Works™ program wwavePDB.


